Abstract
Polygenic scores (PGS) are promising in stratifying individuals based on the genetic susceptibility to complex diseases or traits. However, the accuracy of PGS models, typically trained in European- or East Asian-ancestry populations, tend to perform poorly in other ethnic minority populations and their accuracies have not been evaluated for Native Hawaiians. In particular, for body mass index (BMI) and type-2 diabetes (T2D), Polynesian-ancestry individuals such as Native Hawaiians or Samoans exhibit varied distribution from other continental populations, but are understudied, particularly in the context of PGS. Using BMI and T2D as examples of metabolic traits of importance to Polynesian populations (along with height as a comparison of a similarly highly polygenic trait), here we examine the prediction accuracies of PGS models in a large Native Hawaiian sample from the Multiethnic Cohort with up to 5300 individuals. We find evidence of lowered prediction accuracies for the PGS models in some cases, particularly for height. We also find that using the Native Hawaiian samples as an optimization cohort during training does not consistently improve PGS performance. Moreover, even the best-performing PGS models among Native Hawaiians have lowered prediction accuracy among the subset of individuals most enriched with Polynesian ancestry. Our findings indicate that factors such as admixture histories, sample size, and diversity in GWAS can influence PGS performance for complex traits among Native Hawaiian samples. This study provides an initial survey of PGS performance among Native Hawaiians and exposes the current gaps and challenges associated with improving polygenic prediction models for underrepresented minority populations.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWAS) have identified thousands of genetic variants associated with a plethora of human complex traits and diseases1,2. The success of GWAS has enabled a burgeoning field of post-GWAS analysis, including the computation of polygenic scores (PGS) for predicting the genetic risk for an individual based on their genotypes and available GWAS summary statistics to common diseases or complex traits3,4. On the basis of the GWAS summary statistics, the PGS is calculated as the sum of the trait-associated alleles an individual carries, weighted by the estimated effect sizes of the alleles. Recent developments have focused on extending PGS modeling to include variants and their appropriate weights genome-wide5. Such genome-wide PGS tend to be more efficacious in predicting outcome than a simple approach of selecting only variants that surpassed the genome-wide significance threshold in a GWAS6,7,8.
As the sample sizes of GWAS increased, the resulting PGS has become more effective in risk stratification, early disease detection, and the development of precision medicine9,10,11,12. However, available GWAS data are heavily biased towards European-ancestry individuals13,14, posing a challenge for PGS due to its poor transferability between populations. In particular, PGS trained with European populations exhibit reduced prediction accuracy when applied to non-European populations15,16,17. This poor transferability has been demonstrated for several ethnic minority populations but has not been evaluated for Native Hawaiians, who make up the largest Pacific Islander population in the U.S18.
Native Hawaiians are known to have a higher risk of obesity, diabetes, and cardiovascular disease, among others, both within Hawaiʻi and nationally19,20,21,22,23. They also have the highest mortality rate for several types of cancer compared to other ethnic groups24,25,26. In addition, as consequence of centuries of colonization and globalization, Native Hawaiians are largely admixed27,28,29, with a primary ancestry component descended from Polynesian ancestors (~40%) and recent (within the last 300 years or so) admixtures from European (~30%) and East Asian (~29%) immigrants, among others. These ancestries for Native Hawaiians are correlated with elevated risks of certain metabolic diseases, including body mass index (BMI) and type-2 diabetes (T2D)19,20,21,23. Despite these elevated disease risks, the Native Hawaiian population is largely understudied and underserved27,29. We generally lack the genomic resources and knowledge for this population to reap the benefits of genetic research and genomic medicine27,30,31.
Predictions based on PGS are one area that is under-investigated for Native Hawaiians and can be potentially improved. To date, no systematic evaluation of PGS, particularly for metabolic traits and diseases such as BMI and T2D, have been conducted for the Native Hawaiians. We focused on these traits as they are the most available and because they are closely linked to obesity, diabetes, and cardiovascular disease – diseases that show elevated risks within the Native Hawaiian population22,32,33. Furthermore, T2D was identified as one of the diseases for which the Native Hawaiian communities expressed the most concerns21,34. We additionally included height in our study as a comparison trait, since it is a highly polygenic (as are BMI and T2D) trait for which we have an extensive understanding of the genetic architecture, thus potentially providing a testing ground to evaluate any transferability issue of PGS. Given the underrepresentation of Polynesian ancestries in genomic studies and references, poor transferability of PGS models trained in the largest GWAS dataset is expected as is often observed with other populations and ethnic minorities. On the other hand, it is unclear whether the admixture alleviates some of the transferability issues of PGS at the population level. Because admixture levels vary across individuals within a population, even if admixture alleviates some of the transferability issues, it could create disparity within Native Hawaiian communities depending on an individual’s genomic similarity to the underrepresented Polynesian ancestries. It is thus crucial to evaluate the transferability of PGS in this population and assess any disparities specific to Native Hawaiians in order to begin bridging this gap.
We conducted the present study using data from the Multiethnic Cohort (MEC)35. We leveraged the data of approximately 5300 Native Hawaiian (MEC-NH), as well as populations as proxies for East Asian- and European ancestries (approximately 19,600 Japanese Americans, MEC-J; approximately 8500 White Americans, MEC-W), who were genotyped on the Multi-Ethnic Global Array (MEGA) and Global Diversity Array (GDA) arrays to evaluate the prediction accuracy of PGS for BMI, height, and T2D. We trained PGS models using summary statistics from the largest available consortium GWAS from European (EUR), East Asian (EAS), or multi-ethnic populations for BMI, height, and T2D. Additionally, we assessed the efficacies of published PGS models from the PGS catalog36 in MEC-NH. In each case, we also investigated the model efficacy in subsets of Native Hawaiians with higher estimated Polynesian ancestry. We stress that we utilized this study design to examine health disparities within the Native Hawaiian population, and as a way to evaluate how these models may transfer to other Polynesian-ancestry populations. We used empirical (and potentially noisy) estimates of genetic ancestries to assess the impact on the accuracy of currently available PGS models due to admixture over the last 10–12 generations, which is a product of the colonization of the Hawaiian archipelago by Western countries. Interpretations of these estimates beyond the research context are socially complex and thus should not supplant current practices based on genealogical records and self-reports. These estimates also do not imply any hierarchies or socially meaningful subdivisions in the Native Hawaiian communities.
Results
Overview of the study design
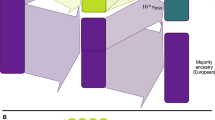
We focused on evaluating the efficacy of PGS models in Native Hawaiians in this study, using the MEC Japanese and White Americans as comparisons. We focused on three traits, BMI, height, and T2D (Supplementary Table 1), and took two different approaches to construct PGS models for evaluation (Fig. 1). We collected from literature the largest East Asian-ancestry (EAS), European-ancestry (EUR), and multi-ancestry (META) meta-analysis GWAS summary statistics for each trait (Supplementary Table 2) to train PGS models. Based on the summary statistics for a particular trait, we used independent, non-overlapping, MEC subsets as reference for linkage disequilibrium (LD) (N = 500) and for PGS optimization (N = 3000), and then validated the PGS model in an additional held-out sample (N = 1000) (Methods, Supplementary Table 3). We trained and optimized PGS models in either EUR-, EAS- or multi-ancestry cohorts, and tested the transferability of the best performing genomic PGS models in Native Hawaiians (Design I in Fig. 1; Supplementary Table 3).
The GWAS summary statistics were downloaded from large consortiums and biobanks (BBJ, UKB + GIANT, BBJ + TWB, and META). Each population-specific GWAS (EAS, EUR) was used to train PGS models with the matching MEC cohort as the LD reference and optimization cohort (MEC-J for EAS, MEC-W for EUR). Multi-ancestry meta-analysis GWAS (META) were used to train PGS with either EAS or EUR populations, each in turn as both LD reference and optimization cohort. In Design I, EAS- or EUR-optimized PGS were validated in held-out MEC-J, MEC-W, and MEC-NH samples. Comparisons of PGS prediction accuracy between MEC-NH and MEC-J or MEC-W provide the metric for transferability. In Design II, PGS models based on EAS or EUR GWAS used MEC-NH for optimization, and the performance in held-out MEC-NH were then compared to the corresponding metric in Design I to assess potential improvement of prediction by PGS. See Supplementary Table 2 for detailed descriptions of the GWAS datasets used for this study and Supplementary Table 3 for a tabular summary of the study design.
Secondly, we explored the potential of leveraging the Native Hawaiian cohort as the optimization cohort to improve the accuracies of PGS models within MEC-NH (Design II in Fig. 1; Supplementary Table 3). Because the Native Hawaiian population, or even the larger Polynesian-ancestry Pacific Islander populations, are generally much smaller in sample sizes compared to the available consortium GWAS from continental populations, it is generally infeasible to amass the sample sizes necessary for a well-powered GWAS that will be informative for PGS construction. We thus investigated whether the MEC-NH could be used for identifying the optimal PGS model to improve upon on the accuracy (hence alleviate the transferability issues) of PGS, even though the GWAS summary statistics were still derived from EAS, EUR, or multi-ancestry meta-analysis.
Reduced prediction accuracy when applying EAS- or EUR-trained PGS to Native Hawaiians in some scenarios
We first assessed the transferability of PGS to the Native Hawaiian population (Design I in Fig. 1). In this case, we identified GWAS summary statistics to build and optimize the PGS model using population-matched cohorts from the MEC, with MEC-J and MEC-W as representatives of the East Asian and European-ancestry cohorts, respectively (i.e. EAS GWAS were optimized using MEC-J; EUR GWAS were optimized using MEC-W; Multi-ancestry GWAS were optimized using either MEC-J or MEC-W separately. MEC-J and MEC-W also provided the respective reference for a pattern of linkage disequilibrium, or LD). We then evaluated the performance of the PGS in held-out MEC-J, MEC-W, and MEC-NH individuals by partial or pseudo R2. Poor transferability is indicated if there is a noticeable drop-off in prediction accuracy when a PGS model optimized in one population (e.g. EAS GWAS optimized in MEC-J) is tested in another population (e.g., MEC-W or MEC-NH validation cohort).
As expected, PGS models optimized in MEC-J or MEC-W showed the highest prediction accuracy in validation cohorts from the same populations (Fig. 2). For instance, for BMI based on GWAS summary statistics from BBJ, the best PGS model optimized in MEC-J achieved the highest partial R2 in held-out MEC-J samples among the validation cohorts tested (partial R2 = 0.059), while the best PGS model based on GWAS summary statistics from GIANT + UKB and optimized in MEC-W achieved the highest partial R2 in held-out MEC-W (partial R2 = 0.088). Moreover, consistent with the expectation of poor transferability, PGS trained in EAS tend to have reduced prediction accuracy in the other continental population. For instance, for BMI, EAS-trained PGS model had reduced partial R2 in MEC-W (0.022 vs. 0.059; one-sided p = 0.041 by bootstrapping), and EUR-trained PGS model performed more poorly in MEC-J (0.043 vs. 0.088; one-sided p = 0.045). PGS models trained from multi-ancestry meta-analysis GWAS sometimes reduced the gap in prediction accuracy between MEC-J and MEC-W, though they do not necessarily have higher population-specific prediction accuracies depending on the traits examined (Fig. 2).
The genomic PGS model with the highest prediction accuracy in optimization cohorts was validated in held-out MEC-J, MEC-W, and MEC-NH cohorts. This figure summarizes the results of analysis in Design I, Fig.1, and details of the model parameter for the best performing PGS model can be found in Supplementary Table 4. The PGS construction method that resulted in best model is represented by circles (for C + T approach) or triangles (for LDpred2 approach). The standard errors for the R2 were calculated using 1000 sets of bootstrap samples. For BMI and height, random 1000 individuals from each of MEC-J, MEC-W, and MEC-NH were used for validation. For T2D, all cases and controls that were not used in training were used for validation: 3313 cases and 6700 controls for MEC-J, 468 cases and 3110 controls for MEC-W, and 389 cases and 549 controls for MEC-NH.
When validating EAS- or EUR-trained PGS in the Native Hawaiian cohort, transferability was not consistently poor, depending on the trait or the GWAS summary statistics used for training (Fig.2). For height, there was a noticeable reduction in prediction accuracy (e.g. for EAS-trained PGS based on the BBJ GWAS, partial R2 = 0.253 in MEC-J to 0.048 for MEC-NH; one-sided p < 0.001). In contrast, EAS- or EUR-trained PGS for BMI showed little drop-off when evaluated in MEC-NH (Fig.2). T2D PGS models showed similar pattern as height, though the overall prediction accuracies (on the liability scale) were lower (Fig.2). For example, EUR-trained PGS based on the DIAMANTE European GWAS performed substantially better in MEC-W than MEC-NH (e.g. pseudo R2 on the liability scale = 0.143 in MEC-W vs. 0.047 in MEC-NH; p < 0.001). EAS-trained PGS based on the DIAMANTE East Asian subset, though still appears to be the most efficacious among MEC-J, were generally poorly predictive across ancestry groups, perhaps reflecting its substantially smaller sample sizes (Fig.2).
PGS optimized in Native Hawaiians did not necessarily improve the PGS transferability
In the absence of a large-scale GWAS in Polynesian-ancestry populations, another possibility to improve the PGS prediction accuracies for Native Hawaiians may be to use the MEC-NH cohort for optimization. In this case, the reference LD sample used is consistent with the ancestry of the GWAS, but MEC-NH individuals are used to identify the best performing PGS model for validation in an additional held-out MEC-NH cohort (Design II, Fig.1; Supplementary Table 3). We then compared PGS models derived in this manner with those optimized using MEC-J and MEC-W (from Design I, shown in Fig. 2). When validated in held-out MEC-NH samples, we observed that in general, PGS models optimized in MEC-NH did not necessarily have better prediction accuracy in the validation cohort (Fig. 3); NH-optimized PGS models largely had similar prediction accuracies compared to those optimized using MEC-J and MEC-W (Fig. 3). The notable exceptions are for BMI when using GWAS summary statistics from UKB + GIANT37(partial R2 = 0.115 vs. 0.081; one-sided p = 0.031 by bootstrapping) and for height when using GWAS summary statistics from BBJ + TWB (partial R2 = 0.118 vs. 0.074; one-sided p = 0.039 by bootstrapping. But note that more accurate models exist using GWAS from UKB + GIANT); in these cases, optimization using MEC-NH provided an improvement in prediction accuracies. These improvements were not observed consistently across GWAS datasets for the same trait or across different traits within the same GWAS dataset. We thus conclude that optimization in MEC-NH did not necessarily lead to improved PGS accuracy compared to using the optimization cohort with consistent ancestry as the cohort generated the GWAS summary statistics and may need to be considered on a case-by-case basis.
For each combination of GWAS-trait PGS models that was previously optimized in MEC-J or MEC-W in Fig.2, the same data was then optimized using MEC-NH samples here (Design II, Fig.1). Previously optimized PGS models and the MEC-NH-optimized models were both validated in the same held-out MEC-NH cohort to evaluate if optimization in MEC-NH would improve the prediction accuracy in Native Hawaiians. Details of the model parameter for the best performing PGS model can be found in Supplementary Table 5. The PGS construction method that resulted in the best model is represented by circles (for C + T approach) or triangles (for LDpred2 approach). The standard errors for the R2 were calculated using 1000 sets of bootstrap samples. For BMI and height, a random subset of 1000 MEC-NH individuals was used for validation. For T2D, 389 cases and 549 controls from MEC-NH were used for validation.
Prediction accuracy of publicly available PGS models for Native Hawaiians
We constructed the evaluated PGS models based on some of the largest and most recent GWAS meta-analysis datasets for BMI, height, and T2D (Supplementary Table 2). In addition, there are a number of published PGS models for these traits in the PGS catalog (URL https://www.pgscatalog.org/), some of which may have used different GWAS datasets or different methodologies for constructing PGS. We thus validated in MEC-NH all of the BMI (N = 102), height (N = 87), and T2D (N = 139) PGS models available as of July 2024, and compared them with our top-performing PGS (Supplementary Data 1).
We found that the PGS developed in this study aligns closely with the best-performing public PGS models for BMI in terms of prediction accuracy, while for T2D there are better performing models in the PGS catalog than what we could develop here (Fig. 4). While not a metabolic trait of emphasis in this study, we found that there exist substantially more accurate PGS models of height for MEC-NH than ones developed and evaluated here (P < 0.001). Notably, some of these top performing models (PGS004779-PGS004782; Fig.4) used an ensemble PGS approach, instead of the two approaches we adopted here, and trained in South Asians38.
PGS models available on the PGS catalog (URL https://www.pgscatalog.org/) as of July 1, 2024 were downloaded for BMI, height, and T2D, and validated in the MEC-NH individuals here (the same held-out validation MEC-NH as in Figs. 2 and 3). Blue points represent the PGS constructed in this study with the highest prediction accuracy in MEC-NH. Circles and triangles indicate that the PGS was derived from C + T and LDPred2 approaches, respectively. Gray points depict PGS from the PGS catalog. Only the top 10 performing PGS models from the PGS catalog were shown; the complete data can be found in Supplementary Data 1. The standard error for the R2 was calculated using 1000 sets of bootstrap samples. For BMI and height, random 1000 MEC-NH individuals were used for validation. For T2D, 389 cases and 549 controls from MEC-NH were used for validation.
Reduced PGS prediction accuracy in the Native Hawaiians most enriched with Polynesian ancestries
At the population level, the accuracy of PGS models in trait predictions or stratification in MEC-NH could, in part, be influenced by admixture. Within the population, Polynesian ancestries exist on a continuum, but because they are the component of ancestries least similar to the ancestries of individuals participating in the GWAS datasets, there may be disparity in PGS prediction accuracy among individuals with different proportions of Polynesian ancestries39. Thus, we explored the accuracy of PGS in subsets of 1000 Native Hawaiians with highest Polynesian ancestries (termed here as PNS), and compared to the 1000 randomly selection individuals. We found that PGS models are less accurate in the PNS subcohort. For instance, the partial R2 for the top performing PGS model for BMI (PGS004734) was 0.118 in randomly selected MEC-NH, but 0.084 when applied in PNS. In fact, across the three traits, the top-performing PGS catalog models we found in MEC-NH generally showed reduced prediction accuracy within the PNS subsets (Fig. 5; P = 8.43 x 10−6 by the binomial sign test). In contrast, the PGS model in this study using the MEC-NH for optimization showed relatively little reduction in performance between randomly selected MEC-NH and PNS (Fig. 6; Supplementary Table 6), although the prediction accuracies were variable compared to validation results previously (Fig. 3), reflecting the smaller samples used here. This may also explain why in some cases the prediction accuracies in randomly selected MEC-NH were lower than that in the same number of PNS individuals, though the differences were generally not significant except for the case of height when using the multi-ethnic meta-analysis GWAS40 (partial R2 = 0.087 and 0.209 in MEC-NH and PNS, respectively, Fig. 6; one-sided P = 0.028). Nevertheless, these findings suggest that while optimizing PGS in MEC-NH may not necessarily lead to improved transferability across Native Hawaiians in general, it could lead to more robust prediction accuracies of PGS model among Native Hawaiians enriched with Polynesian ancestries, and thus may be more applicable to other Polynesian-ancestry populations across the Pacific. Unfortunately, because most of the MEC-NH data were used for optimizing the PGS model, we have fewer individuals enriched with Polynesian ancestry to use as validation, resulting in larger error bars and less stable estimates of the prediction accuracy of the PGS models, and preventing us from evaluating PGS models for T2D as we ended with insufficient number of cases and controls.
Each PGS model from PGS catalog was assessed in validation datasets from MEC-NH, either from randomly selected individuals (white points) or individuals with highest Polynesian ancestry (yellow points). The standard errors for the R2 were calculated using 1000 sets of bootstrap samples. For BMI and height, the validation cohort consisted of randomly selected 1000 MEC-NH or the 1000 individuals with the highest estimated Polynesian ancestry among the entire MEC-NH cohort (see Methods). This was not restricted to the 1000 individuals reserved for validation in Figs. 2–4 as none of the MEC-NH individuals were used in construction of the publicly available PGS models. For T2D, because only 768 individuals could be defined as either a case or control (346 cases, 422 controls) among the 1000 individuals with highest Polynesian ancestry, we compared to 768 individuals (318 cases, 450 controls) randomly selected among all MEC-NH individuals with T2D case/control status.
The PGS models were based on EAS-, EUR- or multi-ancestry GWAS, but using the MEC-NH for optimization. The resulting models were previously validated in 1000 randomly selected MEC-NH individuals in Fig. 3. Here, for fair comparison we validated the same models in a subset of 200 individuals out of the 1000 previously used for validation, but this time comparing 200 randomly selected individuals (white points) to 200 individuals with highest estimated Polynesian ancestries (yellow points). Circles and triangles indicate that the PGS was derived from C + T and LDPred2 approaches, respectively. We did not perform the analysis for T2D due to too few case/control samples, particularly among those with high Polynesian ancestries. The standard errors for the R2 were calculated using 1000 sets of bootstrap samples.
Discussion
In this study, we conducted a systematic assessment of the prediction accuracies and transferability of PGS models for Native Hawaiians. We assessed PGS models constructed in this study as well as models that are publicly available from the PGS catalog. We focused on BMI, height, and T2D, as these are traits or diseases that showed different distributions between Native Hawaiians and other continental populations, and where Polynesian ancestries may be correlated with disease risk19. While we observed that EAS-trained PGS models have reduced prediction accuracy in MEC-W (the MEC cohort representing EUR ancestries) and vice versa, our results revealed that these PGS models at times showed comparable prediction accuracies in the Native Hawaiian cohort, especially for BMI. Empowered by the Native Hawaiian cohort in the Multiethnic Cohort Study (N ~ 5300 with genome-wide genotyping array data), we were also able to evaluate whether using the MEC-NH individuals for optimizing the PGS model could improve PGS prediction accuracies for this population, even though our findings suggested limited improvements. Furthermore, because the Native Hawaiian population displays a continuum of genetic ancestry due to its colonial history, with Polynesian ancestries being the majority ancestry, we also evaluated PGS prediction accuracies among the Native Hawaiians most enriched with Polynesian ancestries (PNS). For publicly available PGS models for BMI, height, and, to a lesser extent, T2D, we did observe a reduction in prediction accuracy when applied to the PNS subcohort. While we cannot discount the influence of potentially shared environments or other social determinants of health, our results suggest that recent admixture with European or East Asian ancestries may be mediating the PGS performance, and these PGS models in general may be even less applicable to the Polynesian-ancestry populations across the Pacific at large.
Across the traits, populations, and GWAS datasets that we examined here for training PGS models for Native Hawaiians, irrespective of the degree of reduction in prediction accuracy, we found that training with European-based GWAS generally produced better-performing PGS models over other GWAS used in training. For instance, the prediction accuracy by partial R2 of height PGS model from the GIANT dataset37 (N > 700,000), optimized in MEC-W, was 0.198 in validation MEC-NH, compared to 0.074 for PGS model derived from BBJ + TWB GWAS data41 (N~260,000) with optimization in MEC-J (one-sided P < 0.001; Fig.2). This is thus reflective of the bias in GWAS, where GWAS for European-ancestry populations are still much larger, and thus more powered and informative for PGS, than for East Asian-ancestry populations. Another hint of this bias can also be observed when PGS was constructed from multi-ancestry meta-analysis data. In this case, the same GWAS dataset was optimized in MEC-W or MEC-J separately, but the resulting PGS still had higher prediction accuracy in held-out MEC-W (partial R2 = 0.286) than MEC-J (0.199; one-sided P = 0.023), respectively. This is potentially due to the European-ancestry populations still representing the majority of the dataset in the multi-ancestry meta-analysis ( ~ 628,000 European-ancestry individuals vs. ~ 179,000 East Asian-ancestry individuals in Sakaue S., et al. 40). Therefore, in addition to increasing sample sizes of future GWAS, increasing representation and diversity of these GWAS will also help improve the PGS accuracy, particularly for a population such as the Native Hawaiians (or African Americans and Latinos) who are not yet a major constituent of multi-ancestry GWAS. This may be particularly important since using the under-represented population for optimization, without fundamentally changing the representation of the GWAS data, did not appear to make meaningful improvements (Fig.3). Future evaluation should focus on methods (e.g.,38,42,43) that can incorporate GWAS summary statistics or PGS models from multiple populations in building a PGS model for Native Hawaiians. Such an approach leveraging GWAS from multiple populations could either integrate GWAS from multiple large-scale continental GWAS or integrate smaller but population-specific GWAS such as those from Native Hawaiians or Samoans44,45, or at least incorporate known population-specific associated variants. However, the latter two approaches may still suffer from noise in the summary statistics due to the smallness of these Polynesian-specific studies. Therefore, a focused effort to expand on education, collaboration, and recruitment with the Polynesian-ancestry communities to enable future GWAS will likely be an essential first step.
While we generally advocate for greater inclusion of diverse populations in future GWAS, which both increase the sample size and the diversity of the GWAS, it could also be difficult to predict the impact on prediction accuracy of the PGS which may be dependent on the trait architecture. For instance, switching from a smaller European-only GWAS to a (marginally) larger multi-ancestry GWAS did not produce PGS models with improve the prediction accuracies for BMI in Native Hawaiians (partial R2 = 0.081 vs. 0.078; Fig. 2). It is therefore important to assess the prediction accuracy empirically, particularly for ethnic minority populations such as the Native Hawaiians.
The variable performance of PGS models in MEC-NH depending on the trait, the GWAS dataset, and the approach for optimization, led to the inconsistent observation of transferability of PGS models between continental populations to MEC-NH. That is, PGS models trained in other continental populations are not necessarily consistently poor-performing in MEC-NH. However, we also observed a greater drop-off in performance, particularly for models from PGS catalog, for the PNS subcohort. Our evaluations thus suggested that continental-level admixture, over the last 300 years or so, may have masked the egregious transferability problems for PGS models for these complex traits in Native Hawaiians and that prediction accuracies likely vary at the individual level within the Native Hawaiian community. We again stress that our use to quantify the proportion of ancestries was solely for the purpose of research use, as a means to evaluate potential disparity in how PGS can be informative within the Native Hawaiian population. These ancestry estimates are potentially noisy, dependent on external references used, and could induce social harm when taken out of the particular research context. Nevertheless, variation in genetic ancestries may influence one’s disease risk at the individual level. Therefore, assessments of one’s polygenic risk profile through approaches46,47 that incorporate genetic ancestries, either at the genomic or local level, ideally without explicit inference of discretely labeled ancestries39, may improve prediction accuracies for Native Hawaiians or for admixed populations in general.
We also found that sample size and diversity in GWAS may complicate the construction and application of PGS in underrepresented populations. This is another obstacle for Native Hawaiians, as the collection and use of genetic data from indigenous populations are fraught with past misuse that led to general mistrust from the community48,49,50. For a population already small in size, the availability of cohorts to optimize these PGS models is also much smaller than that available for other continental populations, which in turn prevents the community in reaping the benefits of genomic medicine. Thus, an ongoing discussion, engagement, and involvement of the community centered around issues of participation in research, genetic data collection, and data sovereignty will be required.
Finally, our results may not generalize to other traits and diseases, and future studies may also focus on outcomes such as cardiovascular diseases for which the Native Hawaiians are also known to be susceptible22,51, or asthma, for which the Native Hawaiian communities expressed explicit concern and should be made as research focus21,34. These future directions will further encourage community participation in research and improve the generalizability of our conclusions here. It is with the multi-pronged strategy of inclusion in genetics research, reduction of biases, and focus on community concerns that we can reduce the disparity in PGS model performance and alleviate health disparity in general.
Material and Methods
GWAS summary statistics and PGS Catalog datasets
To comprehensively evaluate the transferability of PGS for the Native Hawaiian population, an admixed population with substantial East Asian (EAS) and European (EUR) ancestries23, we have utilized EAS and EUR GWAS, as well as multi-ethnic GWAS, summary statistics for training PGS models for BMI, height, and T2D. We collected and curated some of the largest available GWAS summary statistics data from recent publications37,40,41,52,53,54 (Supplementary Table 2). In addition to training PGS models from these latest GWAS, we also obtained and tested all models from the PGS-catalog database (http://www.pgscatalog.org/) related to these three traits, published in the database as of July 1, 2024 (Supplementary Data 1).
In all cases, we downloaded the datasets in GRCh38 coordinates or converted the coordinates to GRCh38 genome build using triple LiftOver55 to ensure alleles are aligned in genomic regions that may have inverted between genome builds. Additionally, we verified the order of reference and alternative alleles in the GWAS summary data by comparing it to the GRCh38 reference genome. We also removed the indels in the GWAS summary statistics data.
Study cohorts and notes on population descriptors
To optimize the PGS models for each trait and to evaluate the prediction accuracy and transferability of each PGS model, we leveraged the individual-level genetic data from the Multiethnic Cohort (MEC) study35. MEC encompasses five major ethnic groups: Japanese Americans, Native Hawaiians, African Americans, Latinos, and Non-Latino Whites. All participants were 45-75 years old at cohort entry, recruited from Hawaii and Los Angeles County. Recruitment was conducted via drivers’ license and voter registration files to ensure broad representation across the population35. Bilingual recruitment materials were provided to address potential language barriers. Within the cohort, up to approximately 70,000 individuals have available genome-wide array data. We utilized the subcohorts genotyped with the Illumina Multi-Ethnic Global Array (MEGA) and Global Diversity Array (GDA) arrays, in total containing 19,677 (MEGA: 5022; GDA: 14,655) Japanese Americans (MEC-J), 11,316 (MEGA: 829; GDA: 10,487) Non-Latino White (MEC-W), and 5388 (MEGA: 4144; GDA: 1244) Native Hawaiians (MEC-NH). MEC-J and MEC-W subcohorts were used as benchmarks for the prediction accuracies of PGS trained and optimized in East Asian and European populations, respectively.
The labels and groupings of these individuals are based on self-report at baseline using terminology that was in practice in the early 1990s when the surveys were distributed to participants. In the survey, participants were asked to provide self-reported ethnic or racial background, marking all that applies, with the options of “Black or African-American”, “Chinese”, “Filipino”, “Hawaiian”, “Japanese (includes Okinawan)”, “Korean”, “Mexican or other Hispanic”, “White or Caucasian”, or “Other”. Most MEC analyses categorize participants into one of the five major racial/ethnic groups mentioned above that were targeted during recruitment, prioritizing group memberships in the following order: “African-American”, “Hawaiian”, “Latino”, “Japanese”, “White”. Thus, if an individual reports “Chinese” and “Hawaiian” in the survey, they would be classified as Native Hawaiian for these analyses. Among the 0.3% of individuals where no response was given for the participant, but existed for the mother and father, a race/ethnicity was created from the parent’s information; otherwise the individual is labeled as “Other.” While the MEC subcohorts used in this study (MEC-J, MEC-W, and MEC-NH) represent multiple levels of self-reported ethnicities, nationality, and genetic ancestries, we continue to use these labels for consistency with other MEC studies and for ease of recognizing and interpreting the historical experience of health disparity for some of these populations, including potential benefits in genomic health through polygenic predictions. We recognize these labels are imperfect proxies for genetic ancestry, but note that our study focuses on a cohort of individuals from a community that emphasize self-identity or genealogical record for group membership (in place of estimated genetic ancestry), who have experienced significant disparity due to social determinants of health, who feel strongly of this identity, and who encompass multiple ancestries on a continuum that would be difficult and unethical to delineate or recategorize.
In limited cases where we examined the potential disparity of PGS prediction within Native Hawaiian communities or evaluated their generalizability to other Polynesian populations, we utilized estimated genetic ancestries. Genetic ancestries for the MEC-NH individuals genotyped on the MEGA array were estimated previously19 through unsupervised ADMIXTURE analysis, combining the MEC with reference samples from 1000 genomes. Additional individuals genotyped on the GDA array here were combined with the MEGA array individuals, and together, the genetic ancestries were re-estimated using the same approach. As before, at K = 4 we found that MEC-NH individuals exhibited known components of ancestries from European, East Asian, and African (interpreted based on 1000 Genomes reference samples), as well as a component of ancestry that is unique to the MEC-NH, presumed to be Polynesian in origin.
Phenotype definitions and quality controls of genetic data
Height and weight were based on participant self-report (in inches or centimeters or in pounds or kilograms, respectively) at the baseline questionnaire, from which BMI was derived. T2D cases were defined based on positive indications on any the three criteria: self-reported T2D diagnosis by a physician or medical professional on any of the five questionnaires (from 1993-2016), the use of medication for treatment of diabetes, or Medicare claim data based on ICD-9 codes (249-250.99) or ICD-10 codes (E11.X)56,57. All other individuals who were still responsive on the last questionnaire (questionnaire 5 between 2012-2016) but were not otherwise deemed a case are considered the controls. The majority of the case definitions are based on Medicare claim data with ICD codes specific to T2D, but self-report definitions based on questionnaires do not differentiate T2D from T1D. There is a chance of mis-reporting in the self-report definition, though it is expected to be small given a minimum age of 45 to enter the cohort.
Individual-level data quality control for individuals genotyped on the MEGA array had been previously done and reported; individuals additionally genotyped on the GDA array followed a similar procedure. Briefly, this includes removing individuals with genotype missingness greater than 5% or mismatched in sex. We also removed SNPs with missingness greater than 2%, those that were duplicated, those that mapped to multiple positions or multiple identifiers when lifted over to hg38, monomorphic or failed Hardy-Weinberg equilibrium test (P < 1e-10). The genotyping data were then imputed genome-wide using TOPMed imputation reference panel (version 2). We did not filter out any self-reported Native Hawaiian individuals on the basis of estimated genetic ancestry, since it is a well-known admixed population with a continuous cline of multiple ancestry components19. Furthermore, it is the community belief that estimated genetic ancestries should not be used as an exclusion criterion for community membership and should not supplant well-established customs of self-identity or genealogical records. Therefore, we included all self-reported Native Hawaiians within the MEC as a single population. At times, we did evaluate the efficacy of PGS models for a subset of MEC-NH with a higher proportion of estimated Polynesian ancestry (the top 1000 individuals, which corresponded to approximately 65%, of Polynesian ancestry; the distribution of admixture proportions can be found in previous publications19,30). We did so both for a better evaluation of using PGS in realizing precision medicine at the individual level and for assessment of the generalizability of PGS models to other Polynesian-ancestry populations.
We randomly subset the individuals into three non-overlapping groups when evaluating PGS models for BMI and height: 500 individuals per population for LD reference, 3000 individuals per population for optimizing the PGS model, and 1000 individuals per population for validating the PGS model (Fig. 1). For T2D, we randomly selected 500 individuals for whom we only have the genetic data as LD reference, and then 800 cases as well as 1500 controls for a total of 2300 individuals per population for optimization. We then used all remaining available cases and controls from each cohort for validation: 3313 cases and 6700 controls for MEC-J, 468 cases and 3110 controls for MEC-W, 389 cases and 549 controls for MEC-NH. For individuals used as LD reference or as validation, we randomly selected individuals who were genotyped on the MEGA array or GDA array, respectively, for each of MEC-J, MEC-W, and MEC-NH. For individuals used in optimizing PGS model, we selected from those genotyped on the MEGA array for MEC-J and MEC-NH and from those genotyped on the GDA array for MEC-W due to the availability of samples.
Polygenic Score development and evaluation
Two commonly used approaches were utilized in this study to construct the PGS model: Clumping and Thresholding (C + T)7,58 and LDpred28. LDpred2 was adopted as comparisons of most genomic PGS construction methods showed similar accuracy, and LDpred2 compared favorably among them59. Moreover, while approaches using genome-wide summary statistics for PGS constructions (such as LDpred2) tend to provide models that outperform those based on the C + T method, there are examples of traits and diseases where C + T method provided more accurate and more simplistic models60. We thus include both approaches in our evaluations.
The C + T method involved clumping SNPs in LD based on r2, and distance (kb) parameters, and thresholding SNPs based on specific thresholds of p-values. On the other hand, LDpred2 employed the Bayesian method to estimate the effects of genetic variants on a specific trait and considered the LD information between genetic variants61. We followed previous studies62 in constructing PGS models from C + T and LDpred2, with the following modifications. For the C + T, we used p-values thresholds of: 0.1, 0.2, 0.5, 0.05, 0.01, 0.005, 1e-3, 5e-4, 1e-4, 5e-5, 1e-5, 5e-6, 1e-6, 5e-7, 1e-7, 5e-8; r2 values (based on the LD reference sample of 500 individuals) of: 0.2, 0.1, 0.01, 0.005, and distance in kilobases (kb) window sizes: 250, 500. Together, we evaluated 128 combinations of parameters to identify the optimal PGS (by partial R2 and pseudo R2 for quantitative and dichotomous traits, respectively; see below) in the optimization sample of 3000 individuals. For LDpred2, we used a grid of values for hyper-parameters/tuning parameters - causal variants (ρ), ℎ2 (the SNP heritability), and sparsity (whether to fit some variant effects to exactly zero) to construct PGS. We used ρ from a sequence of 17 values from 0.01 to 1 on a log-scale, a range of ℎ2 within (0.7, 1, 1.4) × estimated heritability, and a binary sparsity option of either on and off (LDPred2-grid models). In addition, we tested a model assuming infinitesimal causal effects, where each variant is assumed to contribute to disease risk (LDPred2-inf model). In total, we evaluated 103 PRS models using LDPred2.
To evaluate the association of a PGS model in either the optimizing sample or validation sample, we evaluated a regression model using the PGS for each individual as the predictor variable and the trait or disease as the response variable using R (version 4.0.0). Linear models regressed BMI and height against age and age2, and the trait was stratified by sex. We extracted the residuals from the model using the R package stats v3.6.2 and the residuals() function and inverse normalized the residuals using the R package norm v1.0 and the qnorm() function. Finally, we merged the results from males and females. For covariates, we additionally included ten principal components from a Principal Component Analysis (PCA) of the dataset to adjust for the impact due to ancestry and population structure. For T2D, we used logistic regression, with covariates age, sex, and ten principal components. The primary metric of PGS model efficacy is the partial R2 for quantitative traits and pseudo R2 on the liability scale (referred to as the liability R2 throughout our study) for dichotomous traits. Partial R2 was calculated using the R package rsq v2.5 and its function rsq.partial(). Pseudo R2 was calculated using the R package DescTools v0.99.43 and its function PseudoR2(). To obtain the pseudo R2 with only the PGS score effect in the dichotomous trait, we calculate the difference in pseudo R2 between the full model (including PGS) and the partial model (without the PGS score). We then converted the pseudo R2 to the liability scale following the method described in Lee et al. 63, using an estimated lifetime risk of 44.95% and 28.95% for MEC-J and MEC-W, respectively64,65. We could not identify published estimates for lifetime risk for Native Hawaiians, but noted that they are known to have even higher risk of T2D, and thus used 60%. Using alternative values of 50% or 70% did not qualitatively change our results. To calculate the standard errors of or to test the difference in partial or liability R2 for PGS models, we conducted 1000 sets of bootstrap resampling. The transferability of PGS is evaluated based on differences of the partial or liability R2 between two populations or between two PGS models.
Statistics and reproducibility
The statistical analysis and reproducibility of experiments have been discussed throughout the Methods section when describing the study design. In general, samples were randomly selected, stratified by population labels for comparisons, without regard to phenotype distribution or outcome. The study design and sample sizes used for each of LD reference, optimization, and validation are shown in Fig. 1 and Supplementary Table 3. The statistical analysis performed to evaluate significance or assess standard errors are provided in text or in Figure legends. We did not perform replications or have multiple technical replicates, though bootstrap resampling was used to estimate statistical errors.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Genetic data utilized in this study is available on dbGAP (accession number: phs002183.v1.p1). GWAS summary statistics were downloaded from literature; see Supplementary Table 2 for reference. For the downloaded PGS models from the PGS catalog, see Supplementary Data 1. The best newly generated PGS models in this study (shown in Fig. 4) are provided at https://doi.org/10.5281/zenodo.15123413. All other data are available from the corresponding author upon reasonable request.
Code availability
The pipeline used for training, optimizing, and validating the PGS models in this study is publicly available through GitHub: https://github.com/imyingchulo/gprs and https://doi.org/10.5281/zenodo.15117356.
References
Tam, V. et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet 20, 467–484 (2019).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Prim. 1, 59 (2021).
Igo, R. P. J., Kinzy, T. G. & Cooke Bailey, J. N. Genetic risk scores. Curr. Protoc. Hum. Genet 104, e95 (2019).
Goldstein, B. A., Yang, L., Salfati, E. & Assimes, T. L. Contemporary considerations for constructing a genetic risk score: an empirical approach. Genet. Epidemiol. 39, 439–445 (2015).
Wang, Y. et al. Polygenic prediction across populations is influenced by ancestry, genetic architecture, and methodology. Cell Genom 3, 100408 (2023).
Pain, O. et al. Evaluation of polygenic prediction methodology within a reference-standardized framework. PLoS Genet 17, e1009021 (2021).
Choi, S. W., Mak, T. S.-H. & O’Reilly, P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15, 2759–2772 (2020).
Privé, F., Arbel, J. & Vilhjálmsson, B. J. LDpred2: better, faster, stronger. Bioinformatics 36, 5424–5431 (2021).
Lewis, C. M. & Vassos, E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 12, 44 (2020).
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018).
Hoffmann, T. J. et al. Genome-wide association study of prostate-specific antigen levels in 392,522 men identifies new loci and improves cross-ancestry prediction. Nat. Genet. 57, 334–344 (2025).
Hodgson, S. et al. Integrating polygenic risk scores in the prediction of type 2 diabetes risk and subtypes in British Pakistanis and Bangladeshis: A population-based cohort study. PLOS Med. 19, e1003981 (2022).
Fullerton, A. B. & Popejoy, S. M. Genomics is failing on diversity. Nature 161, 164 (2016).
Peterson, R. E. et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 179, 589–603 (2019).
Duncan, L. et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 10, 3328 (2019).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Kachuri, L. et al. Principles and methods for transferring polygenic risk scores across global populations. Nat. Rev. Genet. 25, 8–25 (2024).
Brittany, R., Joyce, K. H. & Paul, J. Detailed look at Native Hawaiian and Other Pacific Islander Groups. https://www.census.gov/library/stories/2023/09/2020-census-dhc-a-nhpi-population.html (2023).
Sun, H. et al. The impact of global and local Polynesian genetic ancestry on complex traits in Native Hawaiians. PLOS Genet. 17, e1009273 (2021).
Moy, K. L., Sallis, J. F., Trinidad, D. R., Ice, C. L. & McEligot, A. J. Health behaviors of native Hawaiian and Pacific Islander adults in California. Asia Pac. J. Public Health 24, 961–969 (2012).
Galinsky, A. M., Zelaya, C. E., Simile, C. & Barnes, P. M. Health conditions and behaviors of Native Hawaiian and Pacific Islander persons in the United States, 2014. Vital Health Stat.3, 1–99 (2017).
Aluli, N. E. et al. Diabetes and cardiovascular risk factors in Native Hawaiians. Hawaii Med J. 68, 152–157 (2009).
Madan, A. et al. More than black and white: differences in predictors of obesity among Native Hawaiian/Pacific Islanders and European Americans. Obesity 20, 1325–1328 (2012).
Native Hawaiian/Other Pacific Islander - The Office of Minority Health. https://minorityhealth.hhs.gov/omh/browse.aspx?lvl=3&lvlid=65.
Haque, A. T. et al. Cancer mortality rates by racial and ethnic groups in the United States, 2018-2020. J. Natl Cancer Inst. 115, 822–830 (2023).
Taparra, K., Qu, V. & Pollom, E. Disparities in Survival and Comorbidity Burden Between Asian and Native Hawaiian and Other Pacific Islander Patients With Cancer. JAMA Netw. Open 5, e2226327 (2022).
Chiang, C. W. K. The opportunities and challenges of integrating population histories into genetic studies for diverse populations: a motivating example from Native Hawaiians. Front. Genet. 12, (2021).
Taparra, K., Miller, R. C. & Deville, C. J. Navigating Native Hawaiian and Pacific Islander cancer disparities from a cultural and historical perspective. JCO Oncol. Pr. 17, 130–134 (2021).
Taparra, K. Pacific Islanders searching for inclusion in medicine. JAMA Health Forum 2, e210153 (2021).
Lin, M. et al. Population-specific reference panels are crucial for genetic analyses: an example of the CREBRF locus in Native Hawaiians. Hum. Mol. Genet. 29, 2275–2284 (2020).
Dinh, B. L. et al. Recombination map tailored to Native Hawaiians may improve robustness of genomic scans for positive selection. Hum Genet. 143, 85–99 (2024).
Aluli, N. E. Prevalence of obesity in a Native Hawaiian population. Am. J. Clin. Nutr. 53, 1556S–1560S (1991).
Maskarinec, G. et al. Diabetes prevalence and body mass index differ by ethnicity: the Multiethnic Cohort. Ethn. Dis. 19, 49–55 (2009).
Galinsky, A. M., Zelaya, C. E., Barnes, P. M. & Simile, C. Selected Health Conditions Among Native Hawaiian and Pacific Islander Adults: United States, 2014. NCHS Data Brief 1–8 (2017).
Kolonel, L. N. et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 151, 346–357 (2000).
Lambert, S. A. et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 53, 420–425 (2021).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Truong, B. et al. Integrative polygenic risk score improves the prediction accuracy of complex traits and diseases. Cell Genom. 4, 100523 (2024).
Ding, Y. et al. Polygenic scoring accuracy varies across the genetic ancestry continuum. Nature 618, 774–781 (2023).
Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021).
Chen, C.-Y. et al. Analysis across Taiwan Biobank, Biobank Japan, and UK Biobank identifies hundreds of novel loci for 36 quantitative traits. Cell Genom. 3, 100436 (2023).
Hoggart, C. J. et al. BridgePRS leverages shared genetic effects across ancestries to increase polygenic risk score portability. Nat. Genet. 56, 180–186 (2024).
Ruan, Y. et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet 54, 573–580 (2022).
Minster, R. L. et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat. Genet. 48, 1049–1054 (2016).
Carlson, J. C. et al. A missense variant in CREBRF is associated with taller stature in Samoans. Am. J. Hum. Biol. 32, e23414 (2020).
Xia, X. et al. A Prism Vote method for individualized risk prediction of traits in genotype data of Multi-population. PLoS Genet. 18, e1010443 (2022).
Marnetto, D. et al. Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals. Nat. Commun. 11, 1628 (2020).
Hudson, M. et al. Rights, interests and expectations: Indigenous perspectives on unrestricted access to genomic data. Nat. Rev. Genet. 21, 377–384 (2020).
Kwan, P. et al. Reservations to Participate in Biospecimen Research among Pacific Islanders. Calif. J. Health Promot. 13, 27–33 (2015).
Garrison, N. A. et al. Genomic research through an indigenous lens: understanding the expectations. Annu. Rev. Genom. Hum. Genet. 20, 495–517 (2019).
Mau, M. K., Sinclair, K., Saito, E. P., Baumhofer, K. N. & Kaholokula, J. K. Cardiometabolic health disparities in native Hawaiians and other Pacific Islanders. Epidemiol. Rev. 31, 113–129 (2009).
Mahajan, A. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 54, 560–572 (2022).
Akiyama, M. et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 49, 1458–1467 (2017).
Akiyama, M. et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat. Commun. 10, 4393 (2019).
Sheng, X. et al. Inverted genomic regions between reference genome builds in humans impact imputation accuracy and decrease the power of association testing. HGG Adv. 4, 100159 (2023).
Polfus, L. M. et al. Genetic discovery and risk characterization in type 2 diabetes across diverse populations. HGG Adv. 2, 100029 (2021).
Maskarinec, G. et al. Association of Obesity and Type 2 Diabetes with Non-Hodgkin Lymphoma: The Multiethnic Cohort. Cancer Epidemiol. Biomarkers Prev. EPI-23-0565 (2023) https://doi.org/10.1158/1055-9965.EPI-23-0565.
Collister, J. A., Liu, X. & Clifton, L. Calculating Polygenic Risk Scores (PRS) in UK Biobank: A Practical Guide for Epidemiologists. Front Genet 13, 818574 (2022).
Ni, G. et al. A comparison of ten polygenic score methods for psychiatric disorders applied across multiple cohorts. Biol. Psychiatry 90, 611–620 (2021).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219–1224 (2018).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet 97, 576–592 (2015).
Jeon, S. et al. Evaluating genomic polygenic risk scores for childhood acute lymphoblastic leukemia in Latinos. Hum. Genet. Genom. Adv. 4, 100239 (2023).
Lee, S. H., Goddard, M. E., Wray, N. R. & Visscher, P. M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 36, 214–224 (2012).
Narayan, K. M. V. Lifetime risk for diabetes mellitus in the United States. JAMA 290, 1884 (2003).
Tomic, D. et al. Lifetime risk, life expectancy, and years of life lost to type 2 diabetes in 23 high-income jurisdictions: a multinational, population-based study. Lancet Diabetes Endocrinol. 10, 795–803 (2022).
Acknowledgements
We would like to thank David Conti and Hailiang Huang for discussions of the analysis and release of pre-publication summary statistics. We would like to thank the Native Hawaiian participants in the Multiethnic Cohort that are involved in this study. The Multiethnic Cohort was funded through grants from the National Cancer Institute (U01CA164973, P01CA168530) and the National Human Genome Research Institute (U01HG007397). We also would like to thank the University of Hawaiʻi Cancer Center’s Native Hawaiian Community Advisory Board for reviewing the study proposal and providing comments on earlier versions of this manuscript. This study is supported by grants from the National Human Genome Research Institute (R01HG011646 to C.W.K.C.) and the Taiwan-USC Postdoctoral Fellowship from Taiwan Education Ministry (to Y.-C.L.). K.T. was funded by the Stanford Cancer Institute (SCI), a National Cancer Institute (NCI)-designated Comprehensive Cancer Center, through the SCI Fellowship Award, SCI Women's Cancer Center Innovation Award, and the SCI Justice, Equity, Diversity, and Inclusion Pilot Grant. The computation for this work was supported by the University of Southern California’s Center for High-Performance Computing (https://hpcc.usc.edu).
Author information
Authors and Affiliations
Contributions
Y.-C.L. performed C + T and LDpred2 analysis, performed statistical analysis, visualized data, and compiled figures and tables for the manuscript. S.J. and T.F.C. implemented and tested the LDpred2 analysis pipeline. H.T., K.A., and B.L.D. further curated and maintained the analysis code and/or performed the evaluation of models from the PGS catalog. M.Y., C.-Y.C., and Y.-F.L. performed BBJ + TWB meta-analysis and provided the data pre-publication. G.M., L.R.W., L.L.M., and C.A.H. generated and provided MEC data. K.T. and N.N. critically reviewed the manuscript and guided the analysis from the Native Hawaiian perspective. C.W.K.C. conceived the study design and acquired funding. Y.-C.L. and C.W.K.C. wrote the manuscript with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
C.-Y. C. is an employee of Biogen. Biogen had no role in the research described in this publication. All other authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Qinqin Huang and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Ani Manichaikul and Johannes Stortz.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lo, YC., Tian, H., Chan, T.F. et al. The accuracy of polygenic score models for BMI and Type II diabetes in the Native Hawaiian population. Commun Biol 8, 651 (2025). https://doi.org/10.1038/s42003-025-08050-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-08050-7